MedFusion Analytics
Our mission: Revolutionize patient care by harnessing the power of multi-modal data
Even with the rise of electronic health records, most radiology diagnostic models today focus on predicting a diagnosis or pathological finding using images alone, often ignoring the valuable contextual information that might be found in other aspects of a patient's record. MedFusion Analytics aims to revolutionize patient care by harnessing this power of multi-modal data. Rather than relying solely on images, we merge images, clinician notes and patient vitals together in an early fusion machine learning model to more precisely diagnose radiological findings. By integrating these complex data, we empower physicians with deeper insights, which ultimately improves patient outcomes, reduces costs, and elevates the overall quality of care.
Our web tool also pushes the boundaries of current diagnostic modeling by offering physicians and medical researchers:
- An intuitive interface - easy-to-use input form for uploading chest X-ray images, free-typing clinical notes, and inputting patient vitals
- Real time predictions in seconds, reducing the time to diagnosis
- A comparative look at our fusion model performance compared to individual models
- Comprehensive documentation to help educate physicians on how we developed our models
With MedFusion Analytics, we strived to develop a holistic diagnostic model that both outperforms traditional computer vision models and encourages physicians to incorporate AI into their practice. We're not just refining healthcare; we're reshaping its future through continuous innovation, setting new benchmarks for precision, and ensuring that our solutions keep evolving to meet the demands of a dynamic medical landscape.
Our Approach
MedFusion Analytics is an early fusion multi-modal model that takes in patient vitals, clinician notes, and chest X-ray images to predict one of five pathological findings: atelectasis, cardiomegaly, lung opacity, pleural effusion, or no findings. With de-identified patient data containing all three data modalities, we first developed state-of-the-art tabular, natural language processing (NLP), or computer vision models, using each data modality individually. We then combined those individual models together, resulting in our pioneering fusion model for radiology diagnosis.
Data
We used the MIMIC-IV database, a large set of retrospectively collected medical records that were de-identified in compliance with HIPAA. This dataset contained clinical notes data for over 200,000 patients, chest X-rays for over 65,000 patients, and other patient data such as vitals signs for over 40,000 patients, all across one or multiple admissions over the duration of the collection period.
To enable appropriate comparison and future fusion developments, we chose to include all patients that had image data, clinical notes, and vital signs (tabular) data. Because the Emergency Department (ED) has a great need for efficient diagnosis and is the highest priority for radiology departments, we focused only on patients in the ED. Based on the remaining data availability and to create a manageable scope for our project, we selected the top four pathological findings – pleural effusion, lung opacity, atelectasis, and cardiomegaly – along with studies with no findings, to create our multiclass classification problem. We preprocessed each data type for quality and consistency and finally split our data into training, validation, and test datasets.
Individual Modeling
For each individual model, we evaluated different frameworks specific to each modality and empirically determined the best performing models across each data type. Our final tabular (patient vitals) model used an XGBoost machine learning algorithm. For the clinical notes, we explored general and domain-specific NLP models and discovered a Bio_ClinicalBERT model performed the best. Likewise, we explored various custom CNNs as well as state-of-the-art computer vision models such as ResNet and DenseNet, and ultimately trained our data using an EfficientNet-B3 model.
Fusion Modeling
There are two predominant fusion modeling approaches today: late fusion and early fusion. The more commonly used approach is late fusion, which takes the predictions from individual models and aggregates them together (via functions such as averaging or majority voting) to output a final prediction. While late fusion models are more popular, they do not allow for interactions between data modalities within the model architecture, which leaves out potentially important synergistic information.
With MedFusion Analytics, we decided to push the boundaries and pursue an early fusion modeling approach, which more holistically combines data or extracted features prior to inputting into a machine learning model to output a final prediction. This approach allows for the fusion models to potentially critically capture interactions between different data modalities, resulting in a superior diagnostic model.
After testing many combinations of features and classifiers, we developed our best early fusion model that is implemented in our MedFusion Analytics web tool. This early fusion model takes the normalized tabular data along with the final layers from our computer vision and NLP models and feeds these concatenated fusion features into a tuned XGBoost classifier.
Evaluation
As is standard in medical modeling, we primarily used AUC, both overall and class-specific, as the main metric to evaluate model performance. Throughout model development, we also recorded and secondarily considered additional metrics, such as precision, recall, and F1 score.
MedFusion vs. Individual Models
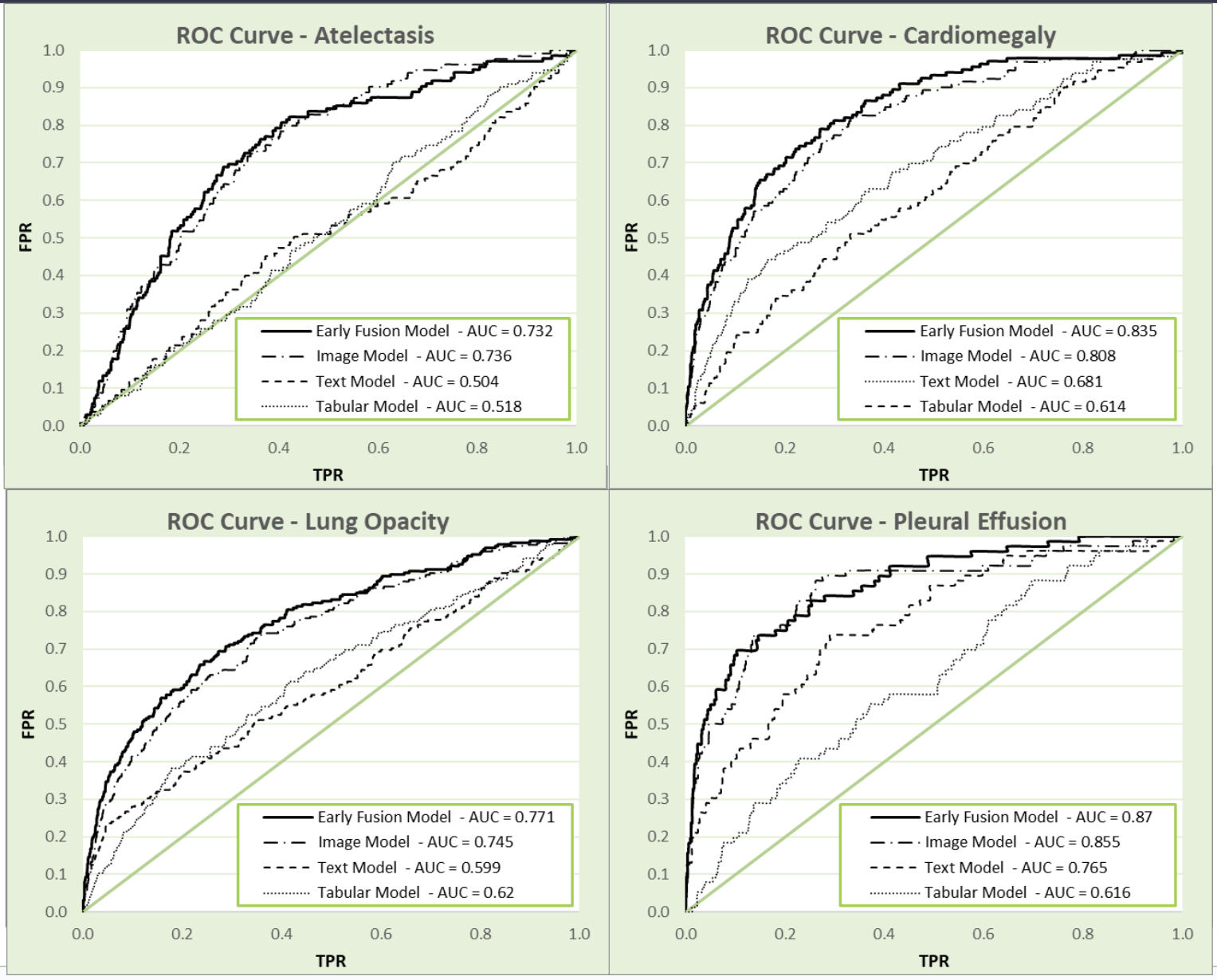
To demonstrate how our fusion model performs as a diagnostic tool, we compared our early fusion model against our best individual models for each pathological finding (Figure 1).
As shown in Figure 1, our early fusion model outperforms both our NLP and tabular models across all four pathological findings, and outperforms our computer vision model in 3 out of the 4 findings (performing similarly in predicting atelectasis, AUC of 0.732 and 0.736 for early fusion and computer vision respectively). Overall, our early fusion AUC of 0.796 outperformed the overall AUC for all three individual models (tabular model AUC = 0.62, NLP model AUC = 0.640, image model AUC = 0.780).
MedFusion vs. Late Fusion Models
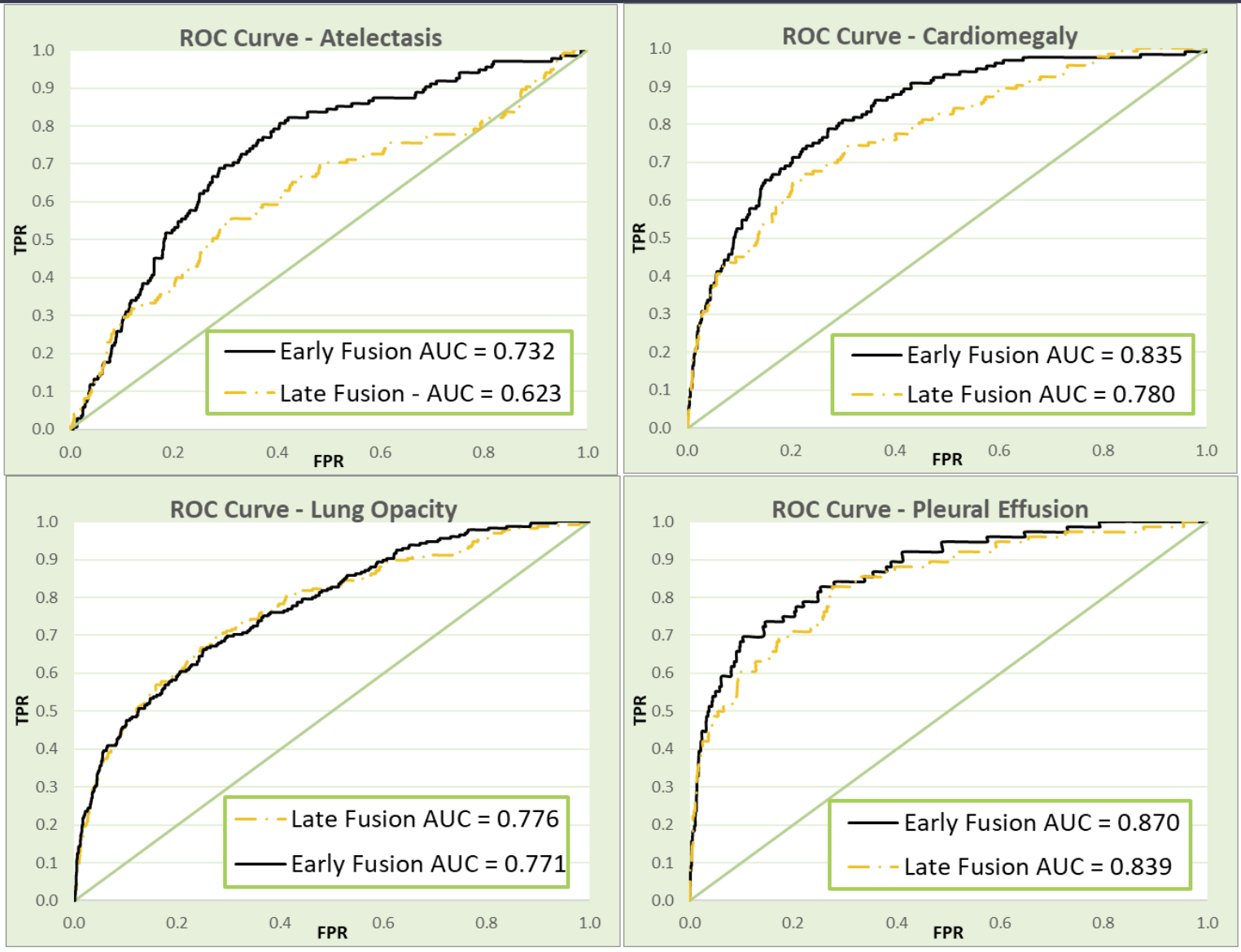
While many fusion models benchmark their performance against models using the individual data modalities, no medically-geared fusion model that we’ve come across has compared their models against other fusion approaches. Because we pioneered an early fusion approach, we also sought to compare our model performance against the more commonly used late fusion models. We generated a late fusion model that averaged the final predictions of all three individual models to compare against our novel early fusion approach (Figure 2).
As shown in Figure 2, our early fusion model clearly outperforms late fusion, notably so in atelectasis and cardiomegaly, which may be due to the more asymptomatic nature of those findings.
Impact
With MedFusion Analytics, we demonstrated the power of taking a more holistic approach to diagnostic AI modeling. We pioneered a top-performing early fusion diagnostic model that we believe will transform how we interact with health data. Our approach empowers physicians to make informed decisions quickly, leading to improved patient outcomes.
We also recognize that AI can be inaccessible to medical professionals due to its “black box” nature and often unfriendly user interface in the clinic. By developing an intuitive web-based tool, we strive to help break down this barrier by encouraging physicians to explore how our model performs using their own data, with real time predictions occurring in seconds. Our users can also explore all of our models and the steps we took to create them, providing the medical community with a valuable education on AI and the future of healthcare diagnostics.
Acknowledgements
We would like to give a huge thanks to our capstone instructors, Korin Reid and Fred Nugen, for their invaluable feedback with this project, as well as our classmates. We would also like to thank the physicians we reached out to who provided us with key insights into the medical space, both from the radiologist and ordering physicians’ perspectives.