Mining the genome for disease mutations
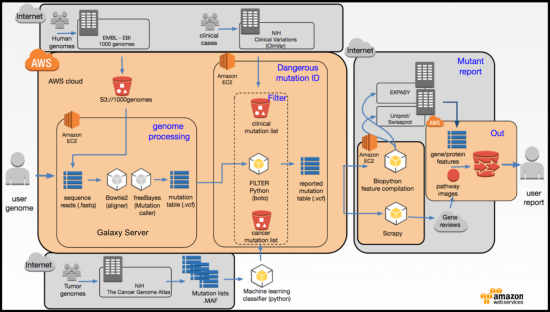
There are at least three major challenges faced by those who want to interpret genome-level mutation data. The first is simply reading and cataloging the 3 billion base pairs of the human genetic variation, this has been undertaken by the 1000 genomes project which hosts data on ~2500 ethnically diverse humans and the Broad Institute who hosts ~68,000 genomes. The datasets are very large and even the major databases have trouble storing and presenting the data. For example, Ensembl recently noted that the smaller of the datasets (1000 genomes) dataset, consisted of 200 billion data points (2500 x 80 million sites) and they had to develop new methods to visualize it. Despite these efforts the data is still very opaque and does not encourage exploration of global patterns; for example, see: 1000 genomes browser, ensemble 1000 genomes, ExAc 68000 genomes browser. Thus the main goal of our visualization is to build an interactive tool to explore the 1000 genomes data in a way that encourages global exploration of the data, allows drill-down to the single gene level, and ultimately links out to gene specific resources on the web.