Multimodal Prediction of Alzheimer's Disease Onset
Problem and Motivation
Alzheimer's Disease (AD) is a complex neurodegenerative disease that severely affects quality of life and is expected to cost the US $1T by 2050. Forty percent of primary care physicians report that they are "never" or "only sometimes" comfortable diagnosing AD. Early detection can result in significantly improved outcomes. In fact, early diagnosis can lower yearly costs by up to twenty percent. We aim to improve health outcomes for Alzheimer's patients by enabling earlier diagnosis of the disease. Contrary to existing approaches which only use MRI data, we use genetic, cognitive, and MRI data to predict the probability of AD onset within the next five years.
Data, Experiments, and Evaluation
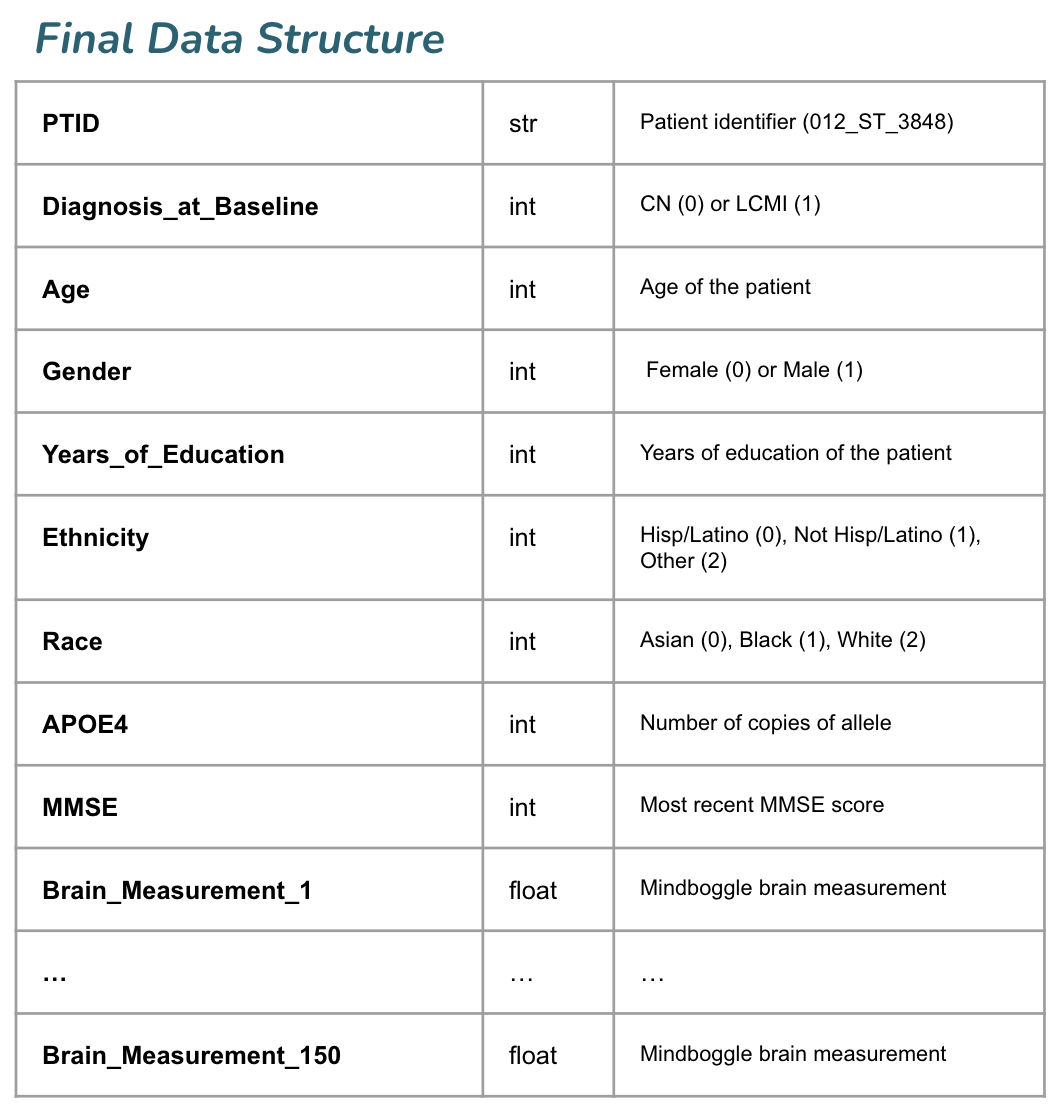
We used data available from the Alzheimer's Disease Neuroimaging Initiative (ADNI) to train our model. Our final dataset includes MRI, genetic, and cognitive data from 606 ADNI participants (this does not include the oversampling we did within the training set to balance classes). Labeled cortical surfaces and labeled cortical and noncortical volumes were extracted from raw 1.5 Tesla T-1 weighted MRI scans using FreeSurfer. ANTs was used for brain volume extraction, segmentation, and registration-based labeling, using the output of the FreeSurfer pipeline as input. Volumes of all labeled regions and thickness of all labeled cortical regions was generated using Mindboggle, using the output of the ANTs pipeline as input. Finally, we used Principal Components Analysis (PCA) to reduce the 2150 Mindboggle measurements to 150 features, which explain roughly 80% of the cumulative variance in the dataset. Below is a sample training record.
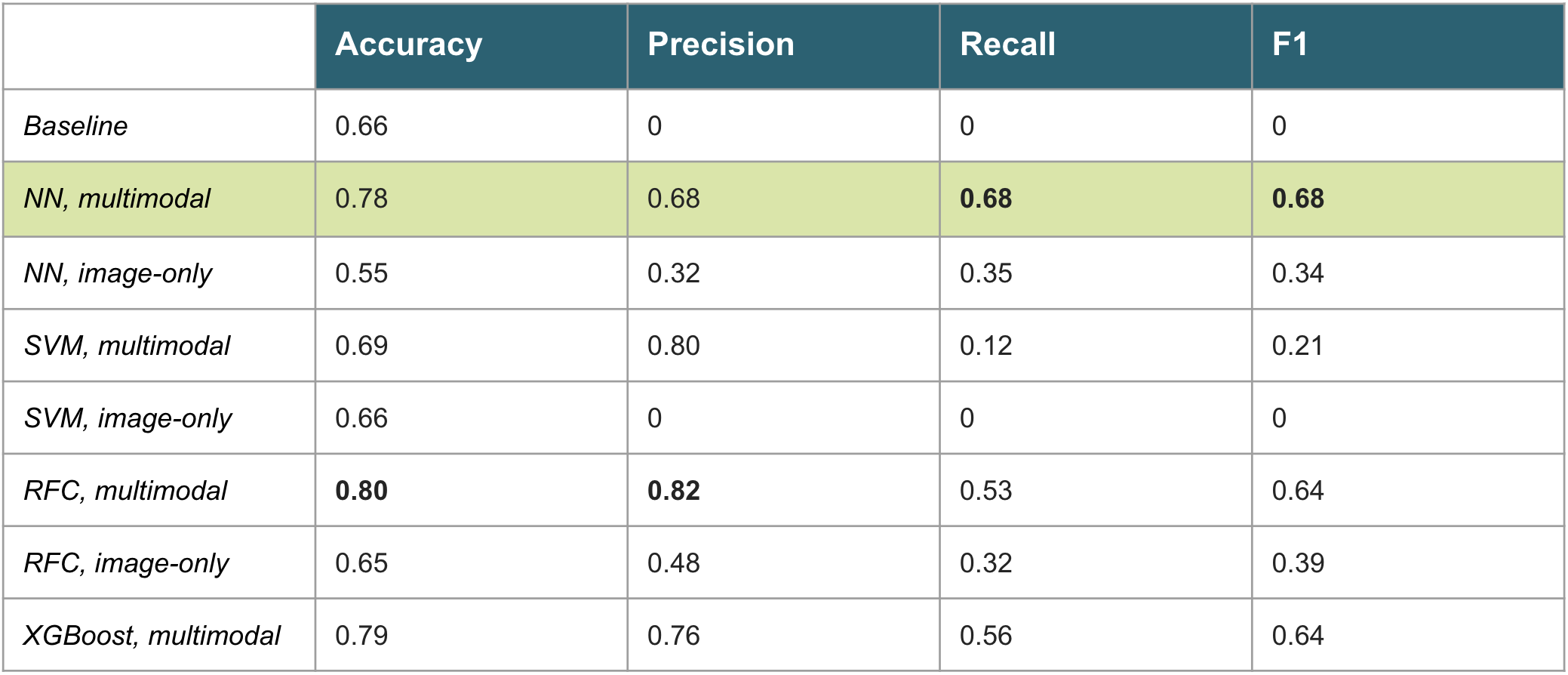
The models we trained include a neural network (NN), support vector machine (SVM), random forest classifier (RFC), and XGBoost. The NN, SVM, and RFC models were trained in the multimodal scenario (see training sample above) as well as image-only scenario (only the 150 Mindboggle measurements with no cognitive or genetic data). The final model we selected for deployment (NN, multimodal) is a fully connected neural network with 2 hidden layers of size 256 and 128. A dropout layer (0.3) follows each hidden layer. We used L1L2 regularization within each hidden layer (1e-3 penalty), a learning rate of 1e-3 with early stopping, and batch size of 16. Finally, we used 5-fold cross validation to assess how the model will generalize. The table below displays test set results for the task of predicting the probability of a given patient developing AD within 5 years.
The baseline represents a model that naively guesses the majority class all the time. Accuracy is the percentage of correct predictions divided by the total size of the test set. Precision is the ratio of true positives to true positives plus false positives. Recall is the ratio of true positives to true positives plus false negatives. F1 is the harmonic mean between precision and recall.
We calculated Shapley values for each feature to evaluate the impact of that feature on the model's predictions. As expected, the patient's diagnosis at baseline has a significant impact on the model's performance (removing this feature reduces accuracy to 0.71). Additionally, more copies of the APOE4 allele and worse MMSE scores push the model's predictions closer to 1 (a diagnosis of AD within the next 5 years). Larger depth of parahippocampal sulci (indicative of impaired working memory) and the perialcarine sulcus (indicative of cortical atrophy) push predictions closer to 1.
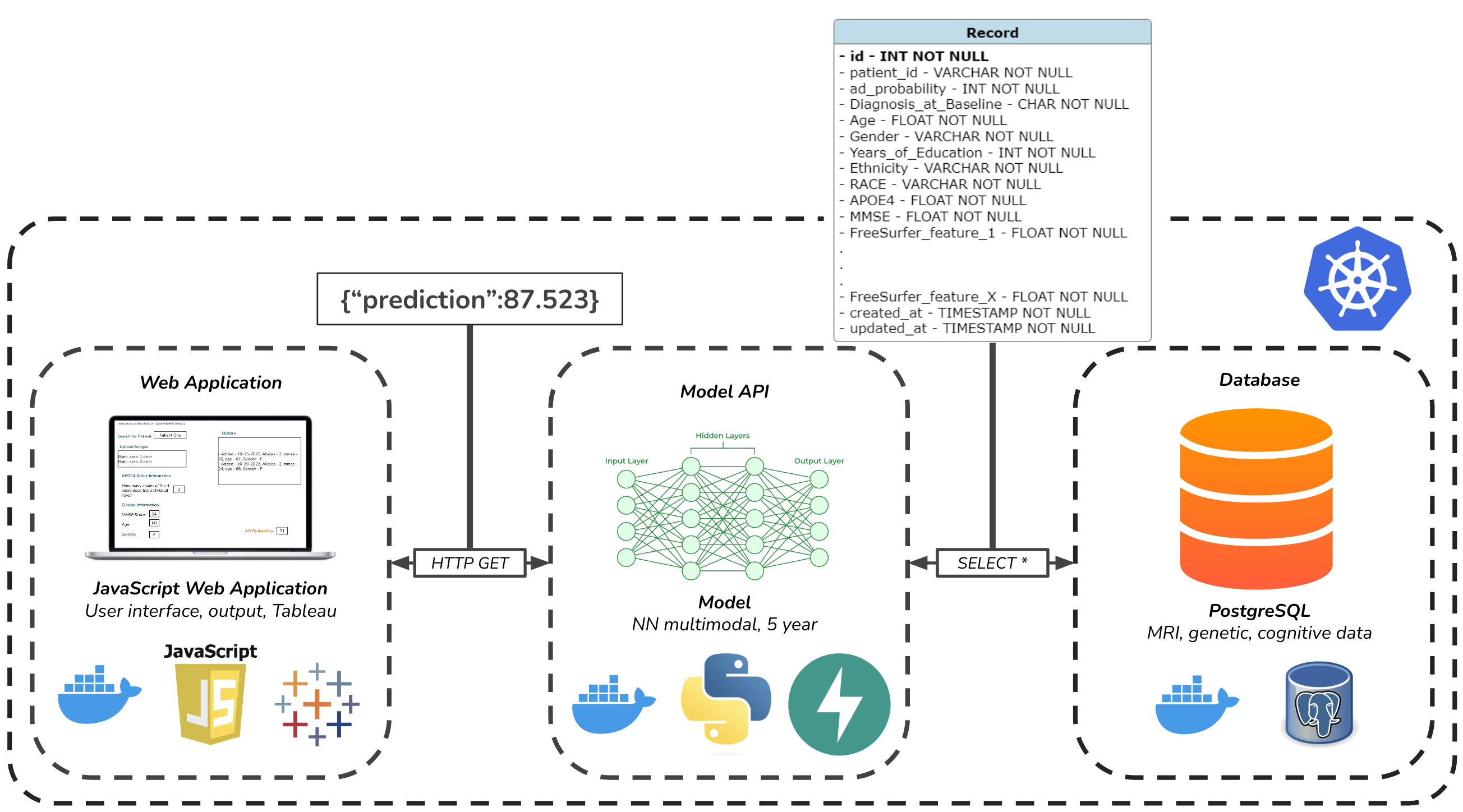
Finally, we developed an API to access the network's predictions, a user interface intended for clinicians, and a database meant to mimic integration with an EHR system. The figure below depicts the architecture of our solution.
Key Learnings and Impact
Incorporation of genetic, cognitive, and clinical data with MRI can significantly improve the performance of models that predict Alzheimer's Disease onset. Accordingly, we believe that our solution has the potential to assist clinicians with earlier Alzheimer's diagnoses and ostensibly reduce the cost of care and improve patient outcomes.
Acknowledgements
We thank Puya Vahabi and Korin Reid for their guidance and support. Additionally, we thank Dr. Nicole Metelski, Ph.D. for her valuable feedback regarding the usability of our application.