LesionLens: A Visual Support Tool for Lesions
Motivation
The Problem
Melanoma is a malignant skin lesion with a substantial global impact, with incidence across the continents ranging from approximately 8,000 in Africa to 150,000 in Europe (World Health Organization, 2020). Early-stage melanomas, along with other malignant skin lesions, are often difficult to differentiate from benign lesions by visual examination alone, even for trained clinicians. Thus, diagnostic processes for skin lesions are inefficient, with unnecessary biopsy rates reaching up to 99.9% (International Skin Imaging Collaboration, n.d.), leading to higher healthcare costs. Additionally, even in areas with access to dermatological care, patients face barriers such as high costs and long wait times. In the United States, the average out-of-pocket cost for an uninsured dermatology appointment is over $150, and wait times for consultations can extend up to 78 days (Walk-in Dermatology, n.d-a, n.d.-b). The health risks associated with delayed or missed melanoma diagnoses emphasize the need to improve visual diagnostic accuracy for skin lesions. The five-year survival rate of this skin cancer ranges from 99% for stage 1 to 30% for stage 4 melanoma (Healthline, 2023). Given the rapid growth of smartphones and mobile health solutions, there is a promising opportunity to enhance access to care through direct-to-consumer tele-consultation services for skin lesions. However, challenges remain, particularly regarding the inconsistent quality of patient-acquired images in telemedicine (International Skin Imaging Collaboration, n.d.). Thus, we are motivated to develop a low-cost, non-invasive, and efficient diagnostic tool for skin lesions.
Our Proposed Solution
Our solution, LesionLens, is a minimum viable product for a web application that offers near-instant visual diagnostic support, providing insights on potential malignancy risk through a machine learning-powered model that takes in user images. Recognizing the limitations of visual assessment, we aim to provide affordable and rapid support to individuals concerned about skin lesions, with a focus on the following target users: high-risk individuals with frequent atypical lesions; people who may avoid healthcare due to cost, convenience, or past negative experiences; and broadly those seeking quick, reliable information about their lesions. While LesionLens does not provide formal diagnoses, it offers diagnostic support and always directs users to consult a dermatologist. LesionLens sets itself apart from existing solutions through its affordability and direct-to-patient accessibility, enabling use without a dermatologist or specialized imaging equipment. We offer clear photo-taking instructions to address common quality issues with patient-acquired images and have designed our model to handle image noise, lighting variations, and positioning inconsistencies.
Data Science Approach
Data Source
The image data we used came from 5 different sources: the BCN_20000 Dataset (Department of Dermatology, Hospital Clínic de Barcelona, n.d.), the HAM10000 Dataset (ViDIR Group, Department of Dermatology, Medical University of Vienna, n.d.), the MSK Dataset (Anonymous, n.d.), and the SIIM-ISIC 2020 Challenge Dataset (International Skin Imaging Collaboration, n.d.). After combining these datasets, we cleaned the resulting data by deduplicating images and removing unlabeled, inconsistently labeled, and inconclusively labeled images. This left us with approximately 20,000 images. The data’s main label was a binary target indicating whether or not a lesion was malignant.
Image Processing
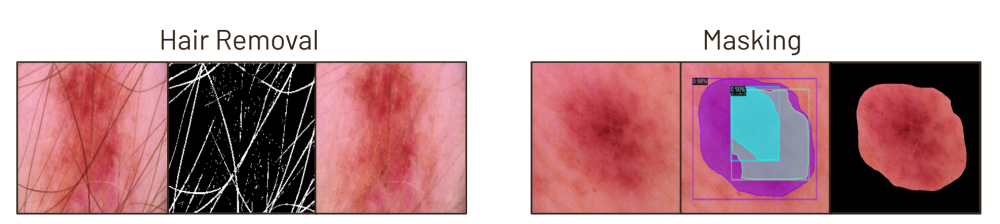
Each image in our dataset underwent several preprocessing steps: resizing, noise removal to eliminate hair and artifacts, masking to isolate the lesion from surrounding skin, and RGB normalization. We experimented with alternative methods, including versions without noise removal and masking. Although these alternatives performed comparably in baseline models, we selected this approach for its ability to effectively isolate lesions from extraneous features that may appear in user photos. Our application applies this preprocessing to user images before feeding it into the classification model.
Class Balancing
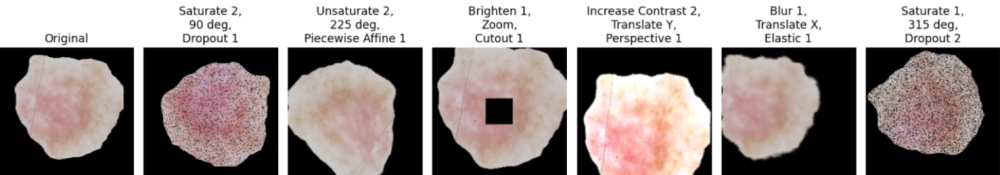
Before training our models, we augmented the training data using layered color, geometric, and warping transformations. This approach aimed to improve our model’s robustness to noise and variations in image quality. Additionally, these augmentations helped balance the dataset by generating synthetic data for underrepresented classes, enhancing performance metrics. We tested other balancing methods, such as undersampling. Although undersampling performed comparably to augmentation, we chose augmentation for its added benefit of handling potential image noise, which aligns with our use case requirements.
Modeling
Our goal was to classify lesions as either malignant or benign. We began by fine-tuning a selection of dense and lightweight transfer learning models, followed by applying ensemble methods to the best-performing models. Based on research supporting effectiveness in lesion classification, we selected and fine-tuned 20 variations of pre-trained image classification models, including MobileNet, ResNet, SENet, InceptionResNet, DenseNet, and NASNetMobile, among others. During training, we focused on optimizing performance and enhancing model generalizability by experimenting with learning rate schedules, optimizers, class weights, dropout layers, and the number of unfrozen layers. Additional design choices, such as early stopping, generator batch size, and data cutoff adjustments, were primarily influenced by memory limitations within our Google Colab environment.
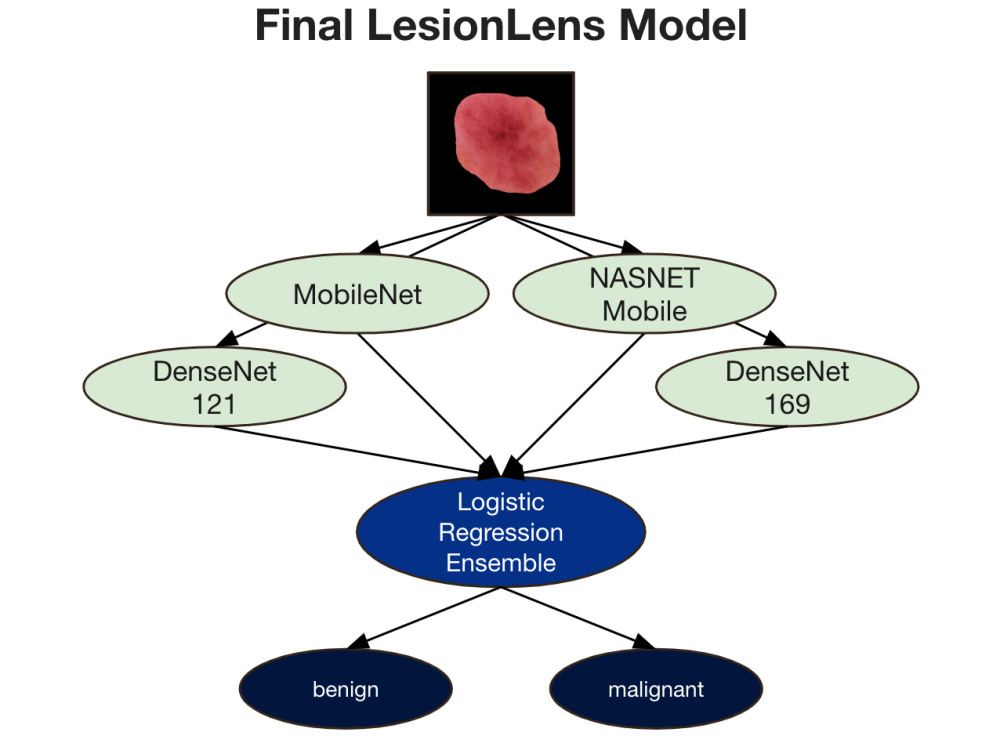
After fine-tuning, we evaluated model performance using accuracy, precision, recall, F1-score, and AUC, selecting the 15 top models out of the original 20 for ensemble experimentation. We initially applied various pruning methods—complementariness, reduced-error, margin distance, and boosting-based pruning—ultimately selecting kappa-error diagram pruning, which performed best with simple averaging across pruned models. This narrowed our ensemble to four models: MobileNet, DenseNet121, DenseNet169, and NASNetMobile. Finally, we experimented with ensemble techniques such as averaging, voting, stacking, and advanced methods, finding logistic regression stacking to yield the best results. Thus, our final model was an ensemble of four fine-tuned models with stacked logistic regression.
Evaluation
Our evaluation benchmarks our final LesionLens model’s performance against both the top-performing external machine learning model on the same dataset (Pham et al., 2020), and the top-performing human expert dermatologist classification on similar data (Haenssle et al., 2018). Our final LesionLens model outperformed human expert classification across all metrics, with around 87% accuracy, precision, recall, and F1 score, and 76% AUC score. Our model demonstrated comparable performance to the best external binary model, with a higher recall but a lower AUC. However, when benchmarking our results, it is important to note that neither the best external model nor the human expert classification reported precision or F1 scores.
| Model | Accuracy | Precision | Recall | F1 Score | AUC Score |
Human Expert Classification | 84.0% | Not reported. | 85.5% | Not reported. | 71.0% |
Best External Model | 88.8% | Not reported. | 83.8% | Not reported. | 88.8% |
LesionLens Final Model | 87.4% | 86.7% | 87.4% | 86.6% | 76.1% |
Key Learnings & Impact
The development of LesionLens underscores several key insights about the potential and limitations of machine learning in dermatological diagnostics. Our primary learning is that accessible, accurate clinical decision support for skin lesions can be achieved through carefully curated image data, model fine-tuning, and ensemble methods. By integrating insights from existing pre-trained image classification models, we successfully created a model capable of distinguishing malignant from benign lesions with performance comparable to the best external machine learning models on similar data.
Our model’s high recall—achieved through strategic choices in ensemble design—addresses a critical healthcare need by identifying high-risk lesions with greater sensitivity than standard visual assessments. This improvement has the potential to reduce missed malignant lesions and, consequently, delay in diagnosis and treatment. Additionally, the direct-to-consumer approach of LesionLens makes it possible to reach underserved populations who face barriers to dermatological care, such as cost and limited access to specialists.
The process also highlighted challenges, including the inconsistent quality of patient-acquired images. We learned that a robust diagnostic model for telemedicine must account for variations in image quality, positioning, and lighting, which required us to design our model and user instructions with real-world application in mind. By offering guidance for image capture and incorporating model adjustments for common visual inconsistencies, we aimed to minimize diagnostic errors arising from these challenges.
Ultimately, LesionLens has demonstrated the feasibility of a low-cost, efficient, and accessible diagnostic tool that can be scaled for broader use. While it is not a substitute for professional medical evaluation, it provides a valuable first step in empowering individuals to seek timely care. By lowering the threshold for accessing reliable skin lesion assessments, LesionLens has the potential to improve early detection outcomes, alleviate unnecessary healthcare costs from over-biopsy, and reduce anxiety for users by providing informative, quick assessments. As mobile health solutions continue to evolve, LesionLens sets a foundation for further advancements in AI-driven tele-dermatology.
Acknowledgements
We would like to express our sincere gratitude to our capstone course instructors, Joyce Shen and Danielle Cummings, for their invaluable guidance, encouragement, and support throughout this project. Their expertise and mentorship were instrumental in helping us navigate the complexities of building a minimum viable product from the ground up. We are also deeply thankful to our fellow 5th Year MIDS students, whose dedication and problem-solving spirit have continually inspired us to push our limits and strive for impactful solutions. This project would not have been possible without the collaborative and motivating environment fostered by our instructors and peers.
References
World Health Organization. (2020). Global Health Observatory: Cancer incidence and mortality. Retrieved from https://gco.iarc.who.int/media/globocan/factsheets/cancers/16-melanoma-of-skin-fact-sheet.pdf
International Skin Imaging Collaboration. (n.d.). About ISIC. Retrieved from https://www.isic-archive.com/mission
Walk-in Dermatology. (n.d.-a). How much does it cost to see a dermatologist without insurance? Retrieved from https://walkindermatology.com/how-much-does-it-cost-to-see-a-dermatologist-without-insurance/
Walk-in Dermatology. (n.d.-b). How long do dermatologist referrals take? Retrieved from https://walkindermatology.com/how-long-do-dermatologist-referrals-take/
Healthline. (2023). What Are the Prognosis and Survival Rates for Melanoma by Stage? Retrieved from https://www.healthline.com/health/melanoma-prognosis-and-survival-rates.
Department of Dermatology, Hospital Clínic de Barcelona. (n.d.). BCN_20000 Dataset.
ViDIR Group, Department of Dermatology, Medical University of Vienna. (n.d.). HAM10000 Dataset. https://doi.org/10.1038/sdata.2018.161
Anonymous. (n.d.). MSK Dataset. Retrieved from https://arxiv.org/abs/1710.05006 and https://arxiv.org/abs/1902.03368
International Skin Imaging Collaboration. (2020). SIIM-ISIC 2020 Challenge Dataset. https://doi.org/10.34970/2020-ds01
Pham, T. C., Hoang, V. D., Tran, C. T., Luu, M. S. K., Mai, D. A., et al. (2020). Improving binary skin cancer classification based on best model selection method combined with optimizing fully connected layers of Deep CNN. 2020 International Conference on Multimedia Analysis and Pattern Recognition (MAPR), Ha Noi, Vietnam. https://doi.org/10.1109/MAPR49794.2020.9237778
Haenssle, H. A., et al. (2018). Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Annals of Oncology, 29(8), 1836–1842. https://doi.org/10.1093/annonc/mdy166