RxReduce: A Medication Deprescribing Agent Framework
Problem & Motivation
Polypharmacy (defined as patients taking 5+ medications) in patients 65+ years of age has tripled over the past few years. Proper medication review with the possibility of deprescribing (elimination of medications from a patient's profile) is a method with which providers can prevent medication interactions, reduce adverse effects, and reduce the complexity of a patient's medication regimen. Due to the complexities of completing the medication review and lack of effective supporting tools, this process is extremely time consuming for providers. As a result, many medications are continued past discharge which could potentially be removed.
Data Sources & Data Science Approach
RxReduce leverages large language models (LLMs) to evaluate patient and make a recommendation along with an explanation with relevant information to enable a clinician to make an informed final decision. Three sources of patient information are evaluated by the system: the patients diagnosis information, inpatient stay information and clinical notes. Unlike the patient diagnosis and inpatient information, a patient may be associated with thousands of clinical notes.
For this work, we focused on the medication class Proton Pump Inhibitors (PPIs) as these are commonly prescribed medication and are frequently continued without supporting indications. In addition there is a robust deprescribing algorithm which we were able to translate into our code. Our test deidentified data sources were acquired through agreement from the Information Commons team at UCSF Medical Center. In addition, for our demonstrations and model evaluation, additional synthetic patient examples were created with were based on actual scenarios we encountered in the deidentified data sources.
Three recommended classes are considered:
- Continue: continue the prescription
- Deprescribe: decrease the amount or frequency of the prescription
- Stop: stop the prescription
For the purposes of evaluation, we had RxReduce provide a single class recommendation along with an explanation containing relevant patient information that led to the decision. During deployment, we will likely encourage providing multiple recommendations where necessary while reporting relevant patient information to empower the clinician to make a recommendation with more complete information.
System Design
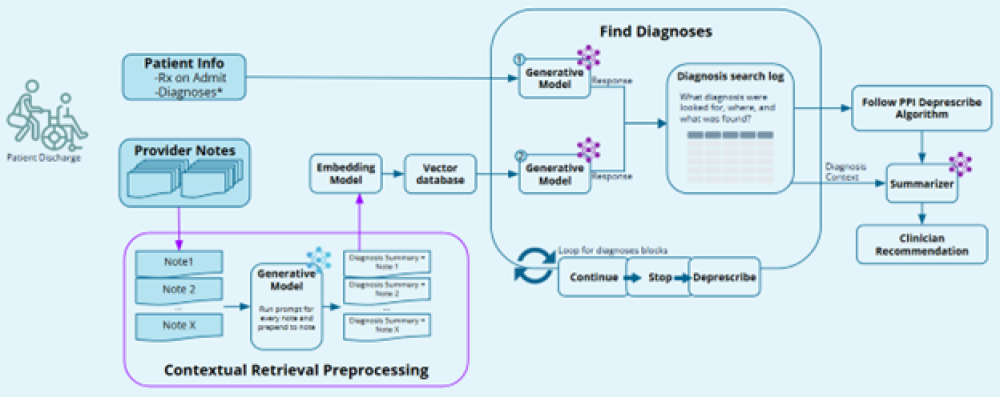
RxReduce uses LLMs to evaluate patient information from multiples data sources sequentially to build a search log containing evaluation findings as well as associated metadata about the data source. The search log containing information regarding relevant patient diagnosis information facilitates the use of the deprescribing algorithm to make the recommendation along with a written explanation citing relevant patient information. Retrieval Augmented Generation (RAG) is used in the case of patient’s clinical notes which may be numerous, while patient diagnosis and inpatient stay data can be considered in their entirety. RxReduce leverages an embedding model that was trained on medical data so that it is exposed to lots of medical terminology.
To make a recommendation decision, the system creates a log as each patient data source is evaluated to determine if it contains diagnosis information associated with each of the recommendation classes (see the deprescribing algorithm linked above). At each step, the system is extracting relevant patient diagnosis information to decide if it is evidence to support a recommendation. As the system conducts the search, results and associated metadata are logged in a json structure. The logged evaluation results are used to make a final recommendation in addition to a summary of the logged findings explaining the recommendation.
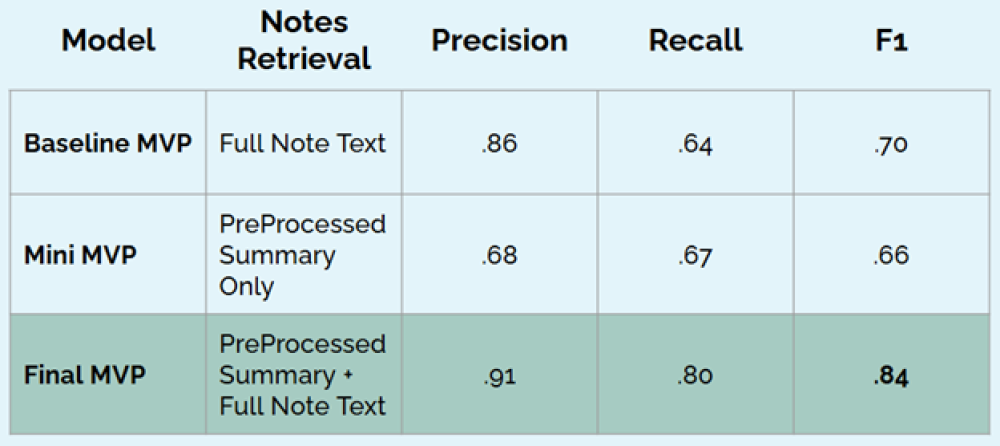
From our evaluations, we found that the clinician notes often contained key patient information needed to make a good recommendation. To improve the ability of the system to evaluate the clinician notes, each patient note was preprocessed by an LLM to generate a summary of the note and determine if it contains information relevant to the medication in question (in this case, a PPI). Then either just the summary, or the summary in addition to the full note were processed by the embedding model and added to the vector database for retrieval. While considering only the summaries resulted in worse results than just considering the full clinician notes, we found the best results came from considering the summary and the full clinician note.
Evaluation
To evaluate RxReduce, we manually evaluated patient cases from the deidentified UCSF data to labels for the best recommendation (continue, stop, or deprescribe) as well as ideal explanations for the decision citing relevant patient information. This is a difficult process with multiple examples where more than one recommendation has reasonable support, and different clinicians might make different recommendations. We created additional synthetic patients to evaluate the system for specific and complicated scenarios. In total, 44 labeled patient examples were considered with six of those being synthetic patients. During a trial, clinician feedback could be collected to rapidly collect many more labeled examples.
RxReduce was evaluated as a classifier (ability to predict the correct recommendation class) in addition to generative metrics focused on response quality using response similarity evaluation. The classification metrics considered included precision, recall and f1 score. System response quality was evaluated with Retrieval Augmented Generation Assessment (RAGAS) faithfulness and relevancy metrics, BERTScore, METEOR, BLEU, and ROUGE.
We focus on the classification metrics here in regards to the contents of the documents in the vector database. Here our baseline MVP is the system when the clinician note texts are considered, the mini-MVP is the system when only the LLM summaries of the clinician notes are considered, the final MVP is the system where the LLM summaries are concatenated with the full clinician note text.
The system performs best as a classifier when considering the LLM summaries concatenated with the full note text.
Key Learnings & Impacts
Our team learned first hand the complexities and time requirements of evaluation of a patients discharge medication list and effort that is required to build a labeled data set. Many of the patients had high note volumes (100+) which we found to be correlated with inpatient stay length. The project demonstrated the viability and power that a LLM powered solution would have in assisting providers with appropriate medication deprescribing recommendations in addition to the location and flagging of the relevant note and diagnosis sections found. We spoke to clinical providers who confirmed the difficulties and time commitment of the discharge medication process and need for better technology to assist them in this crucial step of patient care.
We found success by distributing the search into a sequence to build a system search log which could be referenced to provide relevant contextual information. In practice we would output more than one recommendation where appropriate with relevant explanations and patient information for each. This framework can be used to expand to additional medication classes.[GS1] The system is intended to be agnostic to specific LLM providers although the majority of the work with the system used Llama 3.1 and 3.3 70 billion parameter models. We found that Llama 3.3 expressed more chain of thought reasoning which often resulted in outputting multiple formatted json responses with the final json output being the most refined. In these cases, we chose to extract the final json output. Our evaluations as defined by our label set showed that Llama 3.3 technically performed worse than Llama 3.1. However, although it did not necessarily match our labels, Llama 3.3 often provided reasonable recommendations given the patient information, highlighting the difficulty faced by clinicians. Collecting a larger labeled dataset with clinician feedback would propel further development of the system.
Integrations
RxReduce is deployment ready, conforming to necessary data standards to pull patient data and push results to clinicians. Patient data is queried through a Fast Healthcare Interoperability resource (FHIR). The RxReduce API is called with the necessary patient information to generate a recommendation and associated explanation. The results are pushed to an Electronic Medical Record (EMR) for interaction with the clinician.
Acknowledgements
We extend our gratitude to our instructors, Korin Reid and Ramesh Sarukkai along with appreciation for the feedback and guidance that Mark Butler provided as we worked to optimize our LLM responses. Additionally we thank the UCSF Information Commons team for their curation of the deidentified health data. Finally we thank Matthew Growdon, MD, MPH, Brian L Michaels, PharmD BCPS, Jessica Pourain, MD, FAAP, and Cynthia Fenton, MD for their feedback on the difficulties surrounding providers in the medication discharge process.